Issue Date: September 3, 2012
TB Researchers Revive Old Method In Search For New Drugs
News Channels: Biological SCENE
Keywords: tuberculosis, nonprofits, neglected diseases, pharmaceuticals, antibiotics

After a 40-year drought, the first trickle of new drugs is emerging from the tuberculosis pipeline. They are a welcome addition to a field that has labored with old, inefficient treatments and the emergence of highly resistant strains of the bacteria. But the new pills are just the beginning of what is needed to eradicate TB, a scourge that, according to the World Health Organization (WHO), claimed 1.4 million lives in 2010.
“We should celebrate but not go home,” says Carl F. Nathan, a professor of microbiology and immunology at Weill Cornell Medical College and a leading TB researcher.
Scientists can be forgiven for wanting to go home. For decades the field suffered from a lack of resources and interest. The sequencing of the Mycobacterium tuberculosis (Mtb) genome in the late 1990s reinvigorated researchers, who believed the new data would quickly guide the field toward promising antibacterials. But understanding which targets are important was harder than anyone thought. After spending millions of dollars and chasing dead ends for many years, researchers failed to fill the pipeline substantially.
Committed to learning from the failures, scientists have revamped their approach to TB drug discovery. The field has largely returned to phenotypic screens, which test compounds in whole infected cells, to find starting points for drug discovery campaigns. And with a better understanding of the conditions under which the bug survives in the body, they are trying to conduct smarter drug-screening programs and then work backward to figure out the target of the resulting hits.
Even better, their efforts are more coordinated than before. Nonprofits such as the Bill & Melinda Gates Foundation have convinced big pharma firms and academics to share information in ways not previously seen. Cooperation is expected not only to lower the redundancies inherent in drug discovery but also to help companies quickly decide which projects to scuttle and which ones to prioritize. Largely because of partnerships put together by nonprofit organizations, “a lot more work has happened in the last five years than in the previous 50,” says Manos Perros, head of AstraZeneca’s infectious disease unit.
Mtb is a clever adversary. It sports a thick lipid coat that is hard to penetrate, and it ejects compounds that do squeeze through the protective layer. Even trickier is its ability to hunker down and survive under stress.
Many of the big pharma researchers studying TB cut their teeth on HIV and other infectious diseases. They quickly learn why few new TB drugs have been found. “There have been all these years of low investment and limited innovation, but TB is also infinitely more complex than any infectious disease I’ve come across,” says Nick Cammack, head of GlaxoSmithKline’s Tres Cantos Medicines Development Campus, in Spain, where the company researches neglected diseases.
Combatting TB, scientists say, requires not just one new drug, but three or four drugs that work better than the cocktail of antibiotics now used to treat it. Today’s four-drug regimen of isoniazid, rifampicin, pyrazinamide, and ethambutol takes at least six months to complete. The second-line therapy, taken to fight drug-resistant TB, entails daily injections that carry unpleasant side effects. With neither approach easy to adhere to, development of drug resistance is a constant threat because patients stop taking their medicines once they feel better. At that point, bacteria is still hiding out and can pose a threat months or even years later.
Simply finding more antibiotics is not enough. New antibiotics need to adhere to parameters specific to TB. Each new drug needs to work well with other antibiotics and, because many people with TB also suffer from HIV, with antivirals used to treat HIV. WHO estimates that roughly 10% of TB cases are linked to diabetes, so new TB drugs can’t interfere with diabetes treatments either.
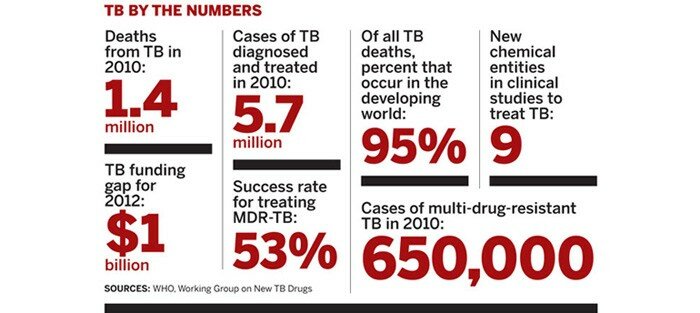
Layer on top of those requirements some practical necessities of TB drugs and the challenge becomes even more daunting. Because TB primarily affects people in the developing world, treatments need to be cheap. Furthermore, new medicines should work within a shorter period than the current six-month standard of care. The lofty goal laid out by the nonprofit Global Alliance for TB Drug Development (TB Alliance) is a three-drug combination that can lick the infection in just 10 days.
Progress is occurring incrementally. The TB community is celebrating two potential new drugs that can be incorporated into the existing regimen, as well as a new three-drug combination that could shorten the time it takes to get rid of the infection.
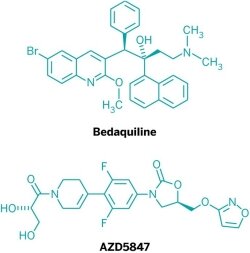
In July, Johnson & Johnson filed a New Drug Application with the Food & Drug Administration for bedaquiline. If approved, the drug would be the first new treatment for TB in more than 40 years and the only one specifically designed to treat multi-drug-resistant TB (MDR-TB). Meanwhile, Otsuka Pharmaceutical has asked European authorities to approve delamanid, a nitrodihydroimidazo-oxazole derivative that is also effective against MDR-TB.
These drugs are “a nice step forward, with a better outcome and more people cured,” says Daniel E. Everitt, senior medical officer at the TB Alliance, which is testing drug combinations that include bedaquiline. The downside, he adds, is that both drug companies have studied the new drugs as additions to the current standard of care, meaning patients will still have to take about seven drugs for at least six months, including daily injections (see page 26).
Another pending drug may address the treatment burden. In July, the TB Alliance unveiled promising Phase II clinical trial results for a new three-drug combination of PA-824, a nitroimidazole the nonprofit is developing; moxifloxacin, an antibiotic marketed by Bayer for other infections; and the TB drug pyrazinamide. In mouse models, the regimen eradicated the bacteria after just three to four months; in human studies, the drug removed 99% of the bacteria in sputum after just two weeks, Everitt says.
The TB Alliance is recruiting patients in Africa for a two-month study that will compare the new three-drug combination with the standard four-drug treatment.
The progress is encouraging, but infectious-disease experts say more and better drugs are needed. “I want to caution against the impression that this problem is about to be solved and we can move on to the next thing. We’ve got to stick with this,” Weill Cornell’s Nathan says. “Resistance is now out there and spreading to all the drugs we have. Introducing one new drug is great, but it isn’t enough.”
Aside from the compounds from J&J, Otsuka, and TB Alliance, the pipeline is largely filled with iterations of existing classes of drugs, namely fluoroquinolones or oxazolidinones, such as AstraZeneca’s AZD5847, or older antibiotics being repurposed to treat TB. They hold promise but are unlikely to cut treatment times.
The paucity of innovation in TB treatment, despite increased R&D funding, is due to a lengthy detour from traditional whole-cell screening into explorations of Mtb’s genome, industry and academic scientists say. Researchers thought the information could be mined for drug discovery. When the Mtb genome was published in 1998, “everybody was enthusiastic and thought, ‘Now we can crack the program really fast,’ ” says Paul Herrling, chairman of the Novartis Institute for Tropical Diseases, the Swiss drug firm’s neglected-disease arm.
The genomics approach involved systematically knocking out genes and determining the effect on Mtb in cultures. The assumption was that if a knockout killed bacteria then the missing gene was essential. The thinking was that the crystal structure of the gene product would be solved and molecules would then be designed to interact with the protein. Academic and industrial labs alike adopted that strategy.
Then in 2007 came a wake-up call. GSK scientists reported the fruits of their target-based screening for new antibiotics, and it wasn’t pretty. Over six years, the scientists screened 67 protein targets against GSK’s vast compound collection, testing up to a half-billion molecules. The effort was “an unprecedented concentration of screening resource for a single therapy area,” the GSK researchers said in their paper (Nat. Rev. Drug Discovery, DOI: 10.1038/nrd2201).
Of the 67 campaigns, only 16 resulted in hits, and just five produced a lead compound. At a price of roughly $1 million per screen and a success rate several orders of magnitude lower than in other therapeutic areas, the company concluded that a target-based approach to finding new antibiotics is unsustainable. When the GSK team analyzed 60 target-based screens conducted outside its labs, it found that not one produced a viable drug candidate.
About 620 genes in the genome of Mtb are believed to be essential for the bacterium’s survival, making the organism particularly laborious to study through a genomic approach. As it turned out, many of the genes essential to the bacteria’s survival in a petri dish are not exactly the same as those critical to the organism living in a human body, Novartis’ Herrling explains.
The findings of the GSK team confirmed what TB drug hunters were experiencing in their own labs as they tried to translate target-based hits into drugs. “People were really beginning to doubt whether that squeeze was worth the juice,” says Mel Spigelman, chief executive officer of the TB Alliance.
With the data showing that target-based screens were a bust, the field was forced to reconsider its methods. Nearly everyone has gone back to whole-cell screens. “You test basically all targets together,” says Koen Andries, a distinguished research fellow at Janssen Pharmaceuticals, J&J’s pharmaceutical arm. “You’re 600 times more efficient.”
Researchers did not have to look far for examples of successes from whole-cell screens: Both the J&J and Otsuka compounds were discovered in them.
J&J stuck with whole-cell screening when many researchers were focused on genetic screening. In the search that led to bedaquiline, J&J screened more than 10,000 molecules against M. smegmatis, which grows faster and is less virulent than Mtb. The screen revealed hits after two days, rather than a week, but the strategy was “a bit of a risk because using a surrogate bacterium means you might miss some compounds which are only active against Mtb,” says Andries, who discovered bedaquiline.
In addition to whole-cell screening, J&J’s chemical library deserves credit for the program’s success, Andries says. “When you try to look for a needle in a haystack, you had better select the right haystack,” he explains. Many academic researchers working on tuberculosis, he notes, don’t have access to high-quality compound collections.
Taking the best hits from the screen, J&J scientists designed 200 diarylquinolones with activity against the bacteria in a petri dish. Three of those compounds were active in mice, and bedaquiline—a molecule with two chiral centers—proved the most potent. Even better, J&J has found that bedaquiline kills both replicating and nonreplicating Mtb.
The downside of whole-cell screening is that the target is unknown and may remain a mystery if multiple targets or pathways are being interrupted. However, J&J successfully worked backward to figure out that bedaquiline targets ATP synthase, an enzyme essential for energy generation.
Whole-cell screening comes with other caveats. A compound that kills bacteria in a petri dish may not work in a mouse model. Scientists have spent the past few years improving the outcomes of screens, trying to mimic the real-world environment of bacteria in the hope of achieving robust hits. They are also pulling in tools from genomics and information gleaned from years of target-based screening to do a better job of translating hits into drug candidates.
One problem that came to light is how researchers were setting up the screens. In 2010, a team of Novartis and academic scientists reported one reason why antibiotics can work well in a petri dish but fail in live animals. Because Mtb grows at a snail’s pace, scientists add artificial carbon sources to the culture media to speed up cell division, Herrling explains. But the extra food causes bacteria to accumulate a debilitating metabolite. Thus, bacterial death that appears to be the effect of an antibiotic would not necessarily occur in a mouse (Nat. Commun., DOI: 10.1038/ncomms1060).
The field is taking a fresh look at other potential pitfalls in its screens. In the human host, sometimes bacteria are replicating and sometimes they are dormant, whereas whole-cell screening typically involves only replicating bacteria. The thinking now is that screening should span multiple environments of the bacteria.
“What is really quite exciting … is this increased understanding that TB exists in different states depending on where it is in the body and what’s happening at the time,” GSK’s Cammack says. For this reason, leading researchers are modifying their phenotypic screens by exposing the cell culture to metabolic or environmental stresses.
For example, Weill Cornell’s Nathan is at the forefront of efforts to unravel the conditions within the host that allow Mtb to reemerge after a period of dormancy: “We try to put TB into a balanced state of dividing and dying, or not dividing at all and not dying at all, during the period of our tests,” he says.
Aided by a grant from the Gates Foundation, GSK is collaborating with Nathan to test compounds on bacteria experiencing environmental stress. Cammack hopes the approach will produce drugs with new mechanisms of action or that can take out Mtb in both its replicating and nonreplicating states. “We might be heading toward more novel approaches to tackling TB,” he says.
TB Alliance’s Spigelman agrees the strategy is worth exploring. “One of the big challenges in TB is we know we can kill 99.9% of the bugs in a really short period of time, but still it takes us months and months to kill that last 0.1%,” he says. If scientists can screen under the conditions in which the bugs hunker down and stop metabolizing, “it might solve the problem of shortening treatment,” he adds.
Others have not given up on the role of the genome in drug discovery. “I still think it’s the road map,” says William R. Bishai, director of the KwaZulu-Natal Research Institute for Tuberculosis & HIV in Durban, South Africa, and codirector of the Center for Tuberculosis Research at Johns Hopkins University School of Medicine. He is convinced that information from the genome will be crucial to understanding the targets of antibiotics discovered through whole-cell screens. “We’ve scratched the surface, but we haven’t really applied what one might call a systems biology approach,” he says.
Clifton E. Barry III, chief of the tuberculosis research section at the National Institute of Allergy & Infectious Diseases (NIAID), agrees that the field would benefit from marrying the best of whole-cell screening, target-based screening, and structure-based drug design. For example, antibiotics researchers at Pfizer used reverse genetics to determine the targets corresponding to hits in a whole-cell screen in Escherichia coli. After solving the target’s crystal structure, the scientists were able to design drugs that were better than the original screening results (Proc. Natl. Acad. Sci. USA, DOI: 10.1073/pnas.0811275106).
“It’s a beautiful story of where we’re at these days,” says Barry, who believes this approach could also work in TB.
New techniques to help researchers understand where a drug or bacteria are in real time could also translate into better drug design. As an example, Weill Cornell’s Nathan points to the use of spatial mass spectrometry to show in real time how an antibiotic is distributed in a tissue of interest. “There’s a great deal of heterogeneity,” Nathan says. “It turns out that when we think we’ve given a therapeutic dose, we’ve given it to part of the lesion but not another part, so of course you’re going to get bacterial resistance.”
As researchers debate the best way forward, funders are promoting efficiency in R&D. When the Gates Foundation provided the latest round of funding for the TB Drug Accelerator, a program seeking to cut TB treatment time to one month, it revamped the accelerator to pool the efforts of seven drug companies, NIAID, the Infectious Disease Research Institute, Weill Cornell, and Texas A&M University.
“What we’re trying to avoid is redundant or futile efforts,” says Ken Duncan, deputy director for global health discovery and translational sciences at the Gates Foundation. In the past, Duncan says, companies worked largely in silos. The foundation’s hope is to convince them to share information in a way that focuses research on the molecules with the greatest promise, regardless of their origin.
A year into the effort, it is clear that sharing information can help researchers prioritize their time. Duncan cites corporate researchers who found a promising hit and were able to compare notes with competitors who came to a dead end with a similar compound. The scientists knew to stop and cut their losses.
Meanwhile, when a whole-cell screen produces a promising hit that has yet to be explored, the compound is shared with the group, and members of the consortium are asked to scour their libraries for related molecules. “You can start to do structure-activity relationships across the companies,” Duncan notes.
NIAID’s Barry says he is seeing companies work together in ways they wouldn’t have in the past. Companies are sharing the structures of their hits and asking traditional competitors to search their libraries for analogs. “That wouldn’t have happened five years ago,” he adds. “There’s definitely a much more open and collaborative feeling.”
The hope is that improved collaboration will allow the TB field to avoid the dead ends that stymied research in the recent past and enable precious research dollars to refill the drug pipeline. “Everything you can do to coordinate research in an attack on this bug is better than going at it alone and making the same mistakes again and again,” Herrling says.
- Chemical & Engineering News
- ISSN 0009-2347
- Copyright © American Chemical Society